As the principal architect of BrainMaps.org, starting with its public launch on May 17, 2005, and ending with my (literally) moving onto new vistas in June, 2009, I can say that it has been an exciting and worthwhile journey. The community of active data contributors, curators, and participants that formed around BrainMaps.org was astounding and exceeded expectations, and it is my hope that this spirit of sharing continues; indeed, that it grows.
What has BrainMaps.org taught us?
First, that an internet-accessible image database of over 60 terabytes of submicron-resolution whole-brain data is feasible with todays technologies. More importantly, however, BrainMaps.org has pointed the way towards the future, towards what lies beyond BrainMaps.org.
What does that future look like?
Imagine, if you will, if the BrainMaps image datasets 1) were imaged at higher resolution, say 40 nm instead of 0.46 micron, 2) had thinner sections, say 40 nm instead of 30 microns, and 3) had all images perfectly aligned into a 3D volume. Then it would be possible to track every neural process and connection in the brain. This is not science fiction. This is the Connectomics revolution that is currently becoming a reality, and it promises to transform the face of neuroscience within the coming decade.
With that in mind, my decision to leave the BrainMaps team in order to join the Connectomics revolution should come as no surprise. I am confident that Ted Jones will maintain the BrainMaps.org site, and hopefully, that it will continue to grow. I am also confident that the Connectomics revolution and the mapping of the mouse brain at nanometer resolution, which will allow us to map out its complete connectivity, will provide a revelation about brain structure and function that is much needed in neuroscience today.
I can still be contacted at brainmaps--at--gmail.com and have more information available at http://Connectomes.org.
best.
Shawn Mikula
Wednesday, January 13, 2010
Tuesday, July 10, 2007
Introducing the Brain Maps API

The Brain Maps API lets you embed Brain Maps in your own web pages with JavaScript. Future versions will enable you to add overlays to brain maps (including markers and polylines) and display shadowed "info windows". The Brain Maps API is a free service, available for any web site that is free to consumers.
Try going to this link to see an example, and once there, try clicking and dragging the image, or clicking on the little tree icon in the upper right. Also, the mouse scroll wheel should zoom you in and out.
Try clicking and dragging the image. Also, the mouse scroll wheel should zoom you in and out.
The code for embedding the Brain Maps API into your own website is simple, requiring just a single line of code:
<iframe src="http://brainmaps.org/ajax-viewer.html?path=http://brainmaps.org/HBP2/c.aethiops/AGM1/AGM1-highres/350/&height=78181&width=92160" width="460" height="267" frameborder="0"></iframe>
It's best to actually download the Brain Maps API and upload the required javascript to your own site, since using just the embedding code above relies on cross-site scripting which is disabled in FireFox.
Additional information is available at http://brainmaps.org/index.php?p=brain-maps-api
Thursday, May 24, 2007
Attack of the Portugese!
- Shawn Mikula

This diagram shows the visitors to BrainMaps.org for the period of April, 2007. The large spike in mid-April, which was larger than the one in February from our press release, was due to a very large influx of visitors from Portugal. We hypothesized that it was due to a Portugese media release, but have never been able to confirm this.

This diagram shows the visitors to BrainMaps.org for the period of April, 2007. The large spike in mid-April, which was larger than the one in February from our press release, was due to a very large influx of visitors from Portugal. We hypothesized that it was due to a Portugese media release, but have never been able to confirm this.
Thursday, March 8, 2007
BrainMaps First Press Release
- Shawn Mikula

The first BrainMaps press release, called "Brain Maps Online", was made on Feb 27, 2007. The effect can be seen in the figure to the right, in which the unique daily visitors to BrainMaps jumped from about 1200 to over 4500 in one day. Now, a week after the initial release, traffic is still more than doubled. Monitoring this effect, it is interesting to note the spread from the initial main press release, to several major online newspapers (i.e., EurekAlert, PhysOrg, ZDNet, ScienceDaily), and then from there to multiple blogs and other outlets. The response from this press release has been nothing short of extraordinary, as numerous individuals emailed us important feedback.

The first BrainMaps press release, called "Brain Maps Online", was made on Feb 27, 2007. The effect can be seen in the figure to the right, in which the unique daily visitors to BrainMaps jumped from about 1200 to over 4500 in one day. Now, a week after the initial release, traffic is still more than doubled. Monitoring this effect, it is interesting to note the spread from the initial main press release, to several major online newspapers (i.e., EurekAlert, PhysOrg, ZDNet, ScienceDaily), and then from there to multiple blogs and other outlets. The response from this press release has been nothing short of extraordinary, as numerous individuals emailed us important feedback.
Saturday, February 24, 2007
BrainMaps.org featured on the cover of NeuroImage
- Shawn Mikula

BrainMaps.org was featured on the March 15, 2007, cover of NeuroImage for the article entitled, "Internet-Enabled High-Resolution Brain Mapping and Virtual Microscopy".
This is only the beginning.
BrainMaps.org is far more than a cool online high resolution brain atlas, it is also a research tool. When used in conjunction with tools like BrainMaps Analyze, it is possible to download image data from brainmaps.org and apply image analysis routines. It is also possible to do 3D reconstructions and morphological analyses. Expect to see upcoming publications emphasizing this point.

BrainMaps.org was featured on the March 15, 2007, cover of NeuroImage for the article entitled, "Internet-Enabled High-Resolution Brain Mapping and Virtual Microscopy".
This is only the beginning.
BrainMaps.org is far more than a cool online high resolution brain atlas, it is also a research tool. When used in conjunction with tools like BrainMaps Analyze, it is possible to download image data from brainmaps.org and apply image analysis routines. It is also possible to do 3D reconstructions and morphological analyses. Expect to see upcoming publications emphasizing this point.
Google Earth for the Brain
- Shawn Mikula
 Users of BrainMaps.org have often described it as a Google Maps for the Brain, which is interesting because we have taken Google Maps as an inspiration and a guide for what mapping the brain should be like. In line with this, one of the developers at BrainMaps.org, Issac Trotts, has released a veritable Google Earth for the Brain, called StackVis. In a nutshell, StackVis is a 3D viewer of neuroanatomical sections. But it is revolutionary in the sense that it permits rapid interactive viewing of arbitrarily large images.
Users of BrainMaps.org have often described it as a Google Maps for the Brain, which is interesting because we have taken Google Maps as an inspiration and a guide for what mapping the brain should be like. In line with this, one of the developers at BrainMaps.org, Issac Trotts, has released a veritable Google Earth for the Brain, called StackVis. In a nutshell, StackVis is a 3D viewer of neuroanatomical sections. But it is revolutionary in the sense that it permits rapid interactive viewing of arbitrarily large images.
Conventional microscopy, electron microscopy, and imaging techniques such as MRI and PET commonly generate large stacks of images of the sectioned brain. In other domains, such as neurophysiology, variables such as space or time are also varied along a stack axis. Digital image sizes have been progressively increasing and in virtual microscopy, it is now common to work with individual image sizes that are several hundred megapixels and several gigabytes in size. The interactive visualization of these high-resolution, multiresolution images in 3D has not been possible, until now. StackVis is a tool for interactive visualization of multiresolution image stacks in 3D.
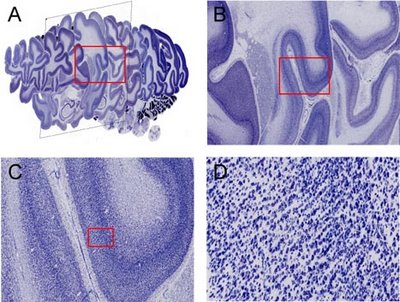 The method, characterized as quad-tree based multiresolution image stack interactive visualization using a texel projection based criterion, relies on accessing and projecting image tiles from multiresolution image stacks in such a way that, from the observer's perspective, image tiles all appear approximately the same size even though they are accessed from different tiers within the images comprising the stack. This method enables efficient navigation of high-resolution image stacks. We implement this method in StackVis, which is a Windows-based, interactive 3D multiresolution image stack visualization system written in C++ and using OpenGL. It is freely available at http://brainmaps.org
The method, characterized as quad-tree based multiresolution image stack interactive visualization using a texel projection based criterion, relies on accessing and projecting image tiles from multiresolution image stacks in such a way that, from the observer's perspective, image tiles all appear approximately the same size even though they are accessed from different tiers within the images comprising the stack. This method enables efficient navigation of high-resolution image stacks. We implement this method in StackVis, which is a Windows-based, interactive 3D multiresolution image stack visualization system written in C++ and using OpenGL. It is freely available at http://brainmaps.org
 Users of BrainMaps.org have often described it as a Google Maps for the Brain, which is interesting because we have taken Google Maps as an inspiration and a guide for what mapping the brain should be like. In line with this, one of the developers at BrainMaps.org, Issac Trotts, has released a veritable Google Earth for the Brain, called StackVis. In a nutshell, StackVis is a 3D viewer of neuroanatomical sections. But it is revolutionary in the sense that it permits rapid interactive viewing of arbitrarily large images.
Users of BrainMaps.org have often described it as a Google Maps for the Brain, which is interesting because we have taken Google Maps as an inspiration and a guide for what mapping the brain should be like. In line with this, one of the developers at BrainMaps.org, Issac Trotts, has released a veritable Google Earth for the Brain, called StackVis. In a nutshell, StackVis is a 3D viewer of neuroanatomical sections. But it is revolutionary in the sense that it permits rapid interactive viewing of arbitrarily large images. Conventional microscopy, electron microscopy, and imaging techniques such as MRI and PET commonly generate large stacks of images of the sectioned brain. In other domains, such as neurophysiology, variables such as space or time are also varied along a stack axis. Digital image sizes have been progressively increasing and in virtual microscopy, it is now common to work with individual image sizes that are several hundred megapixels and several gigabytes in size. The interactive visualization of these high-resolution, multiresolution images in 3D has not been possible, until now. StackVis is a tool for interactive visualization of multiresolution image stacks in 3D.
 The method, characterized as quad-tree based multiresolution image stack interactive visualization using a texel projection based criterion, relies on accessing and projecting image tiles from multiresolution image stacks in such a way that, from the observer's perspective, image tiles all appear approximately the same size even though they are accessed from different tiers within the images comprising the stack. This method enables efficient navigation of high-resolution image stacks. We implement this method in StackVis, which is a Windows-based, interactive 3D multiresolution image stack visualization system written in C++ and using OpenGL. It is freely available at http://brainmaps.org
The method, characterized as quad-tree based multiresolution image stack interactive visualization using a texel projection based criterion, relies on accessing and projecting image tiles from multiresolution image stacks in such a way that, from the observer's perspective, image tiles all appear approximately the same size even though they are accessed from different tiers within the images comprising the stack. This method enables efficient navigation of high-resolution image stacks. We implement this method in StackVis, which is a Windows-based, interactive 3D multiresolution image stack visualization system written in C++ and using OpenGL. It is freely available at http://brainmaps.org
Labels:
3d brain,
brain atlas,
brain mapping,
neurons,
visualization
About this Blog
 |
| Side view of my brain based on MRI. It's not really blue |
Since Blogger does not allow HTML in the 'About Me' section, I thought it good to make a separate post, to let readers know a little about one of the people behind BrainMaps.org. I do realize that this is self-indulgent, but this is the only personal post on this blog, so feel free to stop reading now and skip to other posts.
I was born on the eastern coast of Spain, of Hungarian and Spanish descent, moved to NJ, and then to TX, then to MD for graduate school in neuroscience at Johns Hopkins, and am now in sunny CA at the Univ of Calif, where hopefully I'll remain for awhile.
I work in computational neuroscience and neuroanatomy. I am involved with the BrainMaps Project at the University of California, which involves online high resolution brain mapping.
Here are a few publications to give an idea about my work:
- Mikula S, Manger PR, Jones EG (2007) The Thalamus of the Monotremes: Cyto- and Myeloarchitecture and Chemical Neuroanatomy. Phil. Trans. R. Soc. B. PMID 17229579
- Mikula S, Trotts I, Stone J, Jones EG (2007) Internet-Enabled High-Resolution Brain Mapping and Virtual Microscopy."NeuroImage" 35(1):9-15 PMID 17229579
- Trotts I, Mikula S, Jones EG (2007) Interactive Visualization of Multiresolution Image Stacks in 3D."NeuroImage" PMID 17336095
- Mikula S, Niebur E (2007) Exact Solutions for Rate and Synchrony in Recurrent Networks.
- Mikula S, Fontoura-Costa L, Liu X-B, Jones EG (2007) Particle and Cell Segmentation for Light and Electron Microscopy Using Mathematical Morphology.
- Mikula S, Niebur E (2006) A Novel Method for Visualizing Functional Connectivity using Principal Component Analysis."Int J Neurosci" 116(4):419-29 PMID 16574580
- Mikula S, Niebur E (2005) Rate and Synchrony in Feedforward Networks of Coincidence Detectors: Analytical Solution."Neural Computation" 17(4):881-902 PMID 15829093
- Mikula S, Niebur E (2004) Correlated Inhibitory and Excitatory Inputs to the Coincidence Detector: Analytical Solution."IEEE Transactions on Neural Networks" 15(5):957-62 PMID 15484872
- Mikula S, Niebur E (2003) Synaptic Depression leads to Nonmonotonic Frequency Dependence in the Coincidence Detector."Neural Computation" 15(10):2339-58 PMID 14511524
- Mikula S, Niebur E (2003) The Effects of Input Rate and Synchrony on a Coincidence Detector: Analytical Solution."Neural Computation" 15(3):539-47 PMID 12625330
Labels:
about me,
BrainMaps Team,
publications,
Shawn Mikula
Subscribe to:
Comments (Atom)